Conservation and Genomics of Salmonids: Development of New Tools to Monitor Harvest & Adaptation to Environmental Change

The Problem
Salmonids are increasingly threatened by environmental change, gene flow from hatcheries, and over-harvest in mixed fisheries. For species to persistence in changing environments, e.g. changing climate and infectious disease, it is crucial to conserve genetically-distinct populations adapted to different environments. It has been extremely difficult, until now, to identify adaptively-differentiated populations and the genes underlying the adaptive differences (Luikart et al. 2003; Allendorf et al. 2010).
Over-harvest can drive entire populations to extirpation. So might gene flow from hatcheries that contain populations maladapted to the wild. Accurate, real-time monitoring of harvest and gene flow could prevent over-harvest, and allow monitoring of impacts of gene flow from hatcheries on population performance (Cornuet et al. 1999; Manel et al. 2002). However, until now, too few and uninformative DNA markers (non gene-targeted markers) have been available to enable highly reliable determination of origins of many harvested fish.
Solutions
Recent technological breakthroughs in genomics now make it possible to identify and study adaptive genes and adaptively-differentiated populations by screening thousands of important functional genes from individuals in many populations. This large scale gene screening represents a giant leap forward for science, sustainable harvest, and conservation, as it will allow resolution between stocks at fine spatial scale and the monitoring of adaptive genetic variation. It also allows powerful detection of gene flow (e.g. from hatcheries to wild populations)
Two key advances have made feasible the targeted-sequencing of thousands of functional genes: (1) "Gene capture" arrays (Fig. 2; Mamanova et al. 2010), which allow isolation of many genes from a DNA or tissue sample, and (2) "next generation" DNA sequencing (Eid et al. 2009) which allows rapid analysis of thousands of gene sequences. Together, these techniques now allow us to identify adaptive genes and adaptively-differentiated populations. We propose to apply these molecular genetic technologies, and associated novel statistical methods to identify adaptive gene markers, and to assign individuals to their population of origin (Luikart et al. 2003; Manel et al. 2002; Antao et al. 2008; Schwartz et al. 2009; Olsen et al. 2010; Cosart et al. In review).
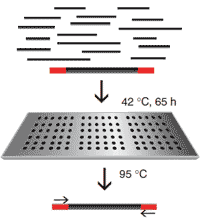 |
|
| Fig. 2. Illustration of gene capture technology that allows targeted-sequencing of 1000s of functional genes. |
|
The Approach
We will use gene capture arrays to isolate and sequence 5,000 functional genes (~20% of all genes) from Pacific salmon. We will sequence wild salmon from many different environments (e.g., high and low temperature spawning populations) in several species, Olsen et al. 2010).
Significance and Expected Outputs
This research will transform salmonid management and conservation by allowing quick and inexpensive assessment of genetic variation in numerous wild stocks and hatchery populations. This project will discover many adaptive gene markers with high inter-population differences allowing for (a) identification of adaptively-differentiated populations, and (b) rapid (real-time) monitoring of harvest and gene flow. This project will generate a 'universal' and species-specific gene-capture arrays and large baseline data sets crucially needed for long-term monitoring of effects of climate change on key salmonid species. From a basic science perspective, this research will advance understanding of the genetic basis of adaptation and how often the same genes underlie similar adaptation in different species. All sequences, gene capture arrays, and baseline data will be made publicly available and published in international scientific journals.
References Cited
- Allendorf, F.W., P. A. Hohenlohe, G. Luikart. 2010. Genomics and the future of conservation. Nature Reviews Genetics, Invited review.
- Antao, T. et al. 2008. LOSITAN: A workbench to detect molecular adaptation…. BMC Bioinformatics, 9:323.
- Cornuet, J-M., S. Piry, and G. Luikart, A. Estoup, and M. Solignac. 1999. New methods employing multilocus genotypes for selecting or excluding populations as origins of individuals. Genetics, 153:1989-2000.
- Cosart, T, et al. Exome-wide DNA capture and sequencing in domestic and wild species. In press.
- Eid, J. et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science, 323:133–138
- Luikart, G. et al. 2003. The power and promise of population genomics: Nature Reviews Genetics, 4:981-994.
- Manel, S. et al. 2002. Detecting wildlife poaching: identifying the origin of individuals using Bayesian assignment tests. Conserv Biology, 16:650-657.
- Mamanova, L. et al. 2010. Target-enrichment strategies for next-generation sequencing. Nature Methods, 7:111-118.
- Ramstad, K.M. et al. 2010. Recent local adaptation of sockeye salmon to glacial spawning habitats. Evolutionary Ecology, 24:391–411.
- Seeb. J. et al. 2010. Next generation transcriptome sequencing and SNP discovery in chum salmon. In review.
