Predicting Effects Of Climate Change On Population Connectivity Using Landscape Genetics And Computer Simulation Modeling

Summary:
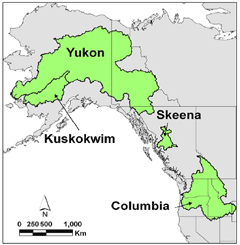
Predicting impacts of landscape and climate change on the connectivity and persistence of natural populations is among the most urgent challenges for society and science. We are developing and applying a landscape genetic analysis framework to advance understanding of how landscape features influence population connectivity and persistence. This combines the use of numerous informative DNA markers, individual-centered landscape resistance modeling, and computer simulations to evaluate the power and reliability of landscape genetic approaches using simulated data and empirical data sets from economically, ecologically, and culturally important species (e.g. salmonids, large mammals, and aquatic insects) from multiple complementary study areas across the northern Rocky Mountains and the Pacific North West USA.
Salmonids are "keystone" species for marine and terrestrial ecosystems (Beamish 2005; Schindler et al. 2003) and can be crucial indicators of ecosystem health in the face of climate change because they are sensitive to warm temperatures (Quinn 2005; Battin et al. 2007). Unfortunately, most native salmonid populations are becoming smaller and more isolated which increases their vulnerability to stochastic events and reduces their ability to adapt through gene flow and demographic rescue. Our research promises to transform landscape and riverscape conservation by establishing well evaluated approaches for combining remote sensing with landscape genomic models that include many DNA markers in functional genes, for multiple species in multiple 'replicate' river systems (figures). This will enable assessment of adaptive potential and associations between genomic and environmental variation at small and large spatial scales crucial for conservation and restoration.
Black bear (Ursus americanus) is an excellent study species for applying landscape genetic approaches because bears are potentially sensitive to habitat change (e.g., warming, drying, intense forest fires or deforestation), and are widely distributed and relatively continuously distributed across diverse landscapes. Individual-centered modeling of different geographic least-cost movement paths identify the landscape features, and their operative scales and relative weighting that most strongly influence gene flow in this species at different spatial and temporal scales. Thus far our studies have produced novel findings that contribute to understandings of black bear ecology, population genetic structure, and gene flow. Using genetic data and individual-based modeling, our study has re-affirmed previous findings (e.g. radio collar data) of the importance of landscape features such as middle elevation and forest cover for black bear movement. Through examining the variation in landscape features within each of multiple study areas, we were able to begin to establish thresholds of variation in landscape features necessary to influence gene flow of black bears. For example, if standard deviation in elevation is greater than c. 300 m, then elevation appears to influence gene flow.
Collaborators:
Salmonids: Fred Allendorf (U Montana), Matt Boyer (MFWP); Clint Muhlfeld (USGS); Robin Waples (NMPS); Jeff Olsen and John Wenburg (USFWS), Shawn Narum (Columbia River Inter-Tribal Fish Commission), Erin Landguth (Research Assistant Prof., U Montana), Mike Schwartz (US Forest Service) and others.
Bears: Fred Allendorf; Neil Anderson & Rick Mace (Montana Department of Fish Wildlife and Parks), Sam Cushman (US Forest Service), Erin Landguth (Research Assistant Prof., U Montana), Kevin McKelvey and Mike Schwartz (US Forest Service), Kate Kendall (USGS), Ruth Short Bull (U. Montana).
Publications:
- Boyer M., G. Luikart, R. F. Leary, and F. W. Allendorf. Comparison of the reliability of non-diagnostic versus diagnostic loci for estimating the proportion of admixture in individuals and populations using a Bayesian approach. In review.
- Landguth, E.L., C.C. Muhlfeld, and G. Luikart. CDFISH: an individual-based, spatially-explicit, landscape genetics simulator for aquatic species in complex riverscapes. In review.
- Landguth, E.L., S.A. Cushman, M. Murphy, and G. Luikart. 2010. Quantifying landscape connectivity: Assessing lag time until barrier signals are detectable. Molecular Ecology Resources. 19:4179–4191.
- Landguth, E.L., S.A. Cushman, M.K. Schwartz, K.S. McKelvey, M. Murphy, and G. Luikart. 2010. Relationships between migration rates and landscape resistance assessed using individual-based simulations. Molecular Ecology Resources. 10:854-862.
- Muhlfeld, C.C. J.J. Giersch, F.R Hauer, G.T. Pederson, G. Luikart, Douglas P. Peterson, Christopher C. Downs, Daniel B. Fagre. 2011. Climate change links fate of glaciers and a rare alpine invertebrate. Climate Change Letters. In press.
- Schwartz, M.K., G. Luikart, K.S. McKelvey, and S. Cushman. 2009. Landscape genomics: a brief perspective. Chapter 19 in S.A. Cushman and F. Huettman (eds). Spatial Complexity, Informatics and Animal Conservation, Springer, Tokyo.
- Short Bull, R.A, R. Mace, S.A. Cushman, E.L Landguth, T. Chilton, K. Kendall, M.K. Schwartz, K.S. McKelvey, F.W. Allendorf, and G. Luikart. 2011. Why replication is important in landscape genetics: Case of the American black bear in the Rocky Mountains. Molecular Ecology. 6:1092–1107.
